
Plotting rjuliabugs draws using bayesplot package
Source:vignettes/using_bayesplot.Rmd
using_bayesplot.RmdOverview
This article focuses on plotting parameter estimates from MCMC draws
obtained using the rjuliabugs::juliaBUGS() sampler. We do
not cover MCMC diagnostics or alternative visualization tools and
packages, although these could be easily adapted, since all plots here
operate on the params array extracted from the
rjuliabugs object. The visualizations shown are based on
those presented in the original vignette by Jonah Gabry and Martin
Modrák. We strongly recommend reading their article: https://mc-stan.org/bayesplot/articles/visual-mcmc-diagnostics.html.
Much of the code and several of the plots here are directly inspired by
their examples.
To begin, we load the necessary models to reproduce the examples:
Example
As previously mentioned the goal of this vignette is exclusively to
show how to use and present some of the visualizations tools from
samples obtained from the sampler from rjuliabugs.
Therefore we will assume that the rjuliabugs model object
is already fitted and named as `rjuliabugs_fit``. For a full walkthrough
until this moment, please refer to the article named Get
Started until the section where we generate the posterior
draws from the sampler
Inspect the params object.
The rjuliabugs is a S3 object which
contains all the information and data used in the sampler. This
includes, from the the the name of identify the model object in the
Julia environment, the mcmc configuration
parameters to the posterior draws named as params which by
default is given by an 3D numeric array with respective dimensions being
(iterations chains parameters). The choice of array as the default is
due to its compatibility with most of other packages in R
enviroment to work with posterior samplers. However, the converstion to
other types to be comptabilite to other packages is possible through the
functions as_rvar, as_mcmc and
as_draws which can be called over the model object itself
(as_rvar(rjuliabugs)) or the rjuliabugs$params
object (as_rvar(juliabugs$params). See the reference
section for a complete documentation of the examples.
At first we need to extract the posterior samples, to do it so we
will use the rjuliabugs::extract function:
pars_names <- c("alpha0","alpha1","alpha2","alpha12","sigma")
posterior_samples <- rjuliabugs::extract(rjuliabugs_fit,pars = pars_names)Verifying the dimensions of the posterior_samples object
we would have
dim(posterior_samples)
#> [1] 2000 4 5we have a total of 2000 iterations, 4 chains and 5 parameters that were stored.
Posterior uncertainty intervals
Central credible intervals from the posterior distribution can be
visualized using the mcmc_intervals function.
mcmc_intervals
color_scheme_set("red")
mcmc_intervals(posterior_samples, pars = pars_names)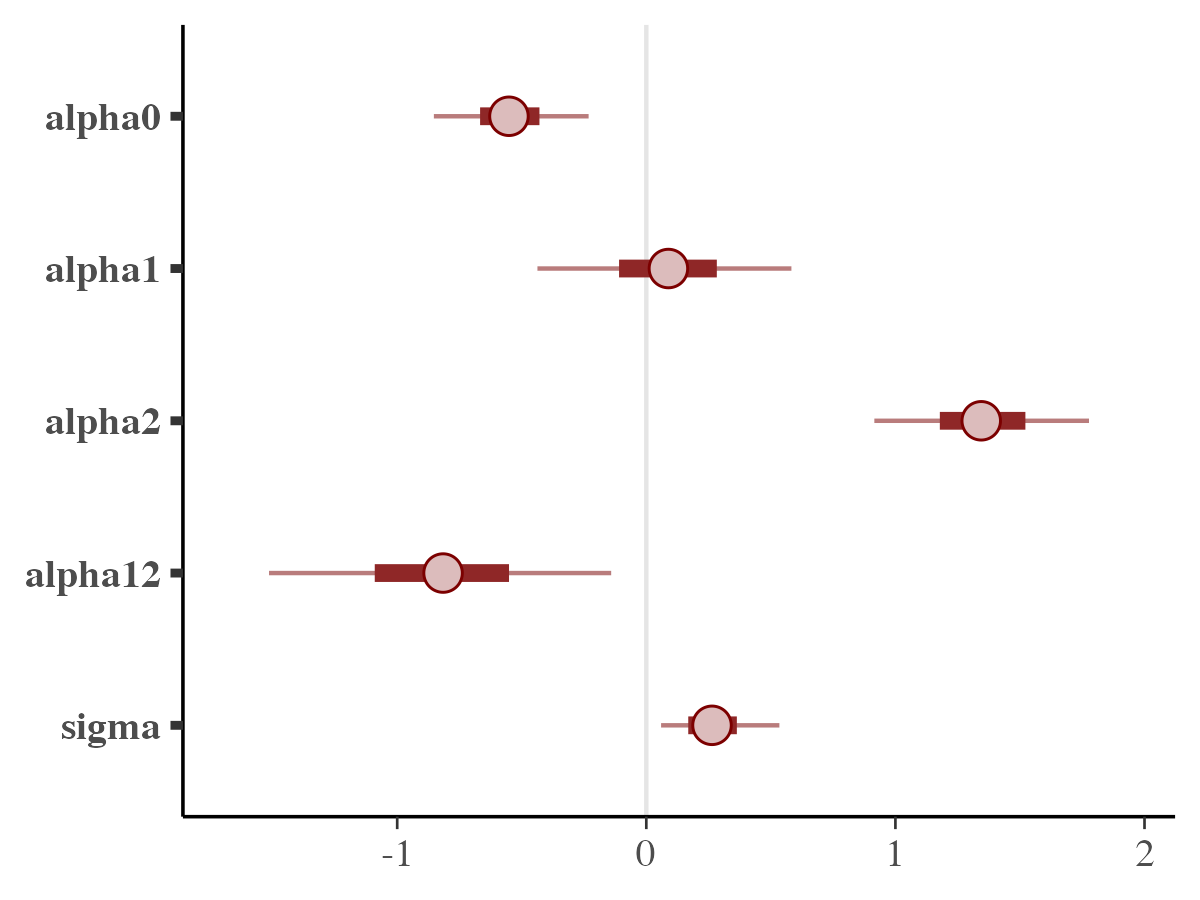
By default, the plot displays 50% credible intervals as bold lines
and 90% intervals as thinner outer segments. These settings can be
adjusted using the prob and prob_outer
parameters, respectively. The plotted points represent the posterior
medians; if preferred, the point_est option allows
switching to posterior means or suppressing point estimates
entirely.
To represent uncertainty with shaded regions beneath the posterior
density estimates, the mcmc_areas function can be
employed.
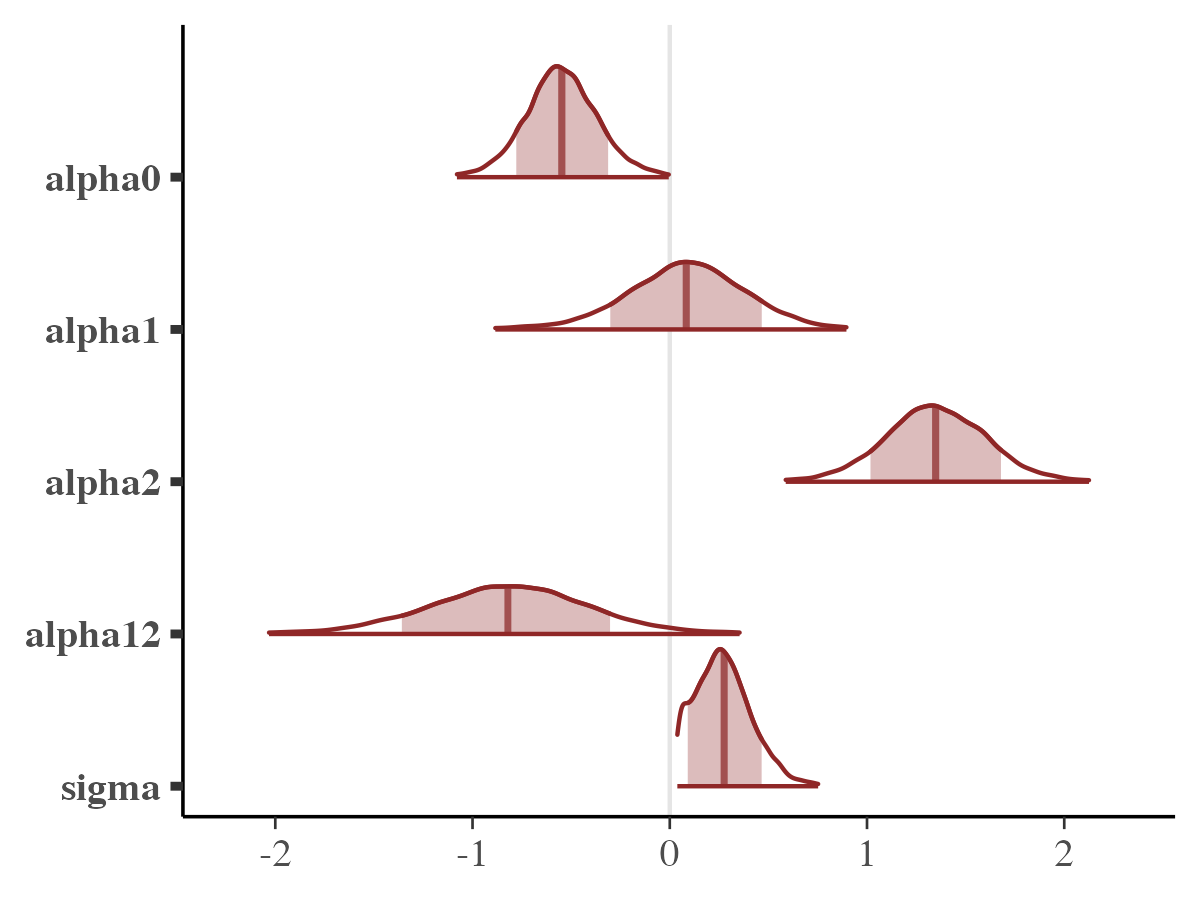
mcmc_intervals
mcmc_areas(
posterior_samples,
pars = pars_names,
prob = 0.8, # 80% intervals
prob_outer = 0.99, # 99%
point_est = "mean"
)
mcmc_areas_ridges
We can also generate additional plots with by calling
mcmc_areas_ridges
mcmc_areas_ridges(
posterior_samples,
pars = pars_names,
prob = 0.8, # 80% intervals
prob_outer = 0.99, # 99%
point_est = "mean"
)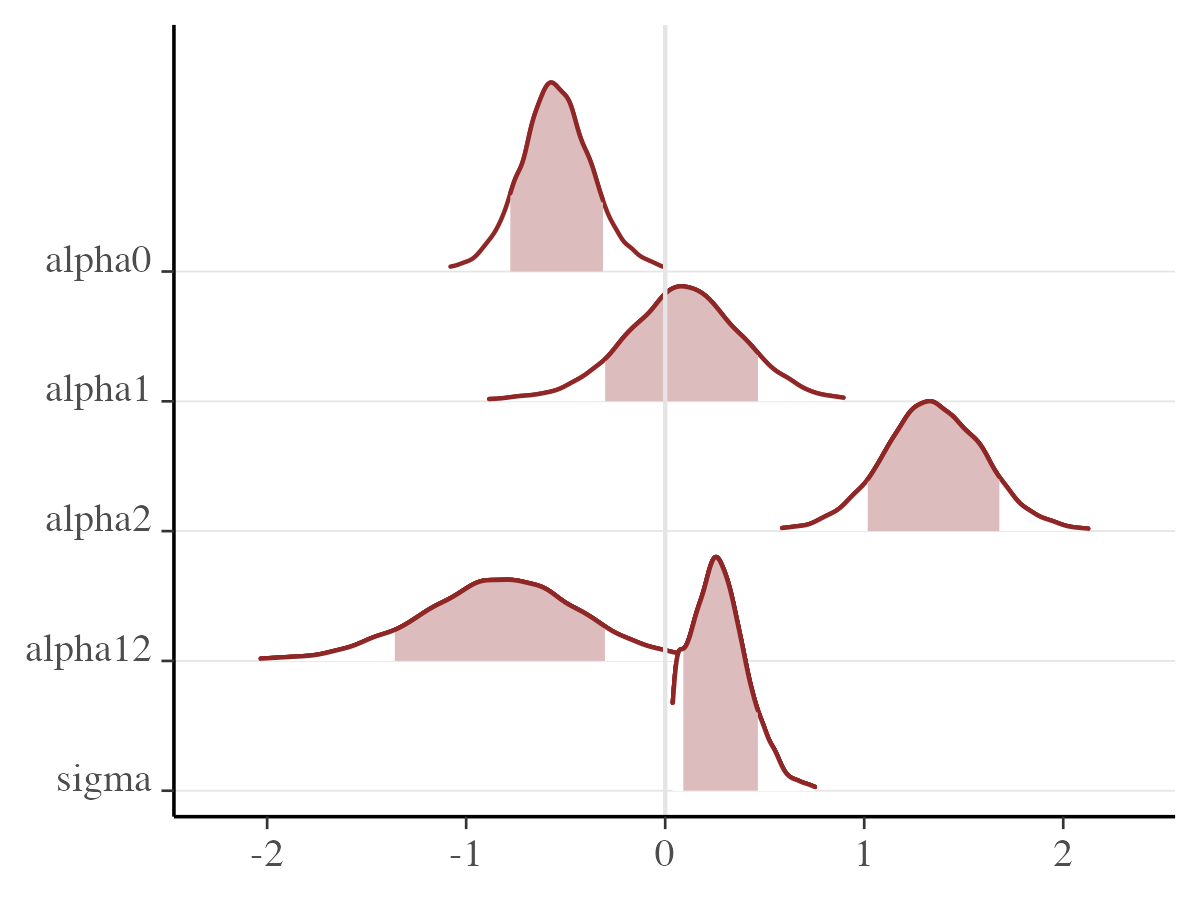 ## Univariate marginal posterior distributions
## Univariate marginal posterior distributions
The bayesplot package includes functions for visualizing
marginal posterior distributions through histograms or kernel density
estimates, either by merging all Markov chains or displaying them
individually.
mcmc_hist
The mcmc_hist function plots marginal posterior
distributions (combining all chains):
color_scheme_set("blue")
mcmc_hist(posterior_samples, pars = c("alpha0", "sigma"))
mcmc_hist_by_chain
To visualize individual histograms for each of the four Markov
chains, the mcmc_hist_by_chain function can be used; it
generates separate facets for each chain within the plot.
color_scheme_set("darkgreen")
mcmc_hist_by_chain(posterior_samples, pars = c("alpha0", "sigma"))If we are interested in also display a transformation for display any
of the histograms, this is possible by setting the
transformations argument.
color_scheme_set("darkgreen")
mcmc_hist_by_chain(posterior_samples, pars = c("alpha0", "sigma"),
transformations = list("sigma" = "log"))Many of the plotting functions for MCMC draws also include a
transformations argument to apply transformations to the
parameters before plotting.
mcmc_dens
The mcmc_dens function works similarly to
mcmc_hist, but it displays kernel density estimates in
place of histograms. This include a version of
mcmc_dens_by_chain.
color_scheme_set("brightblue")
mcmc_dens(posterior_samples, pars = c("alpha0", "sigma"))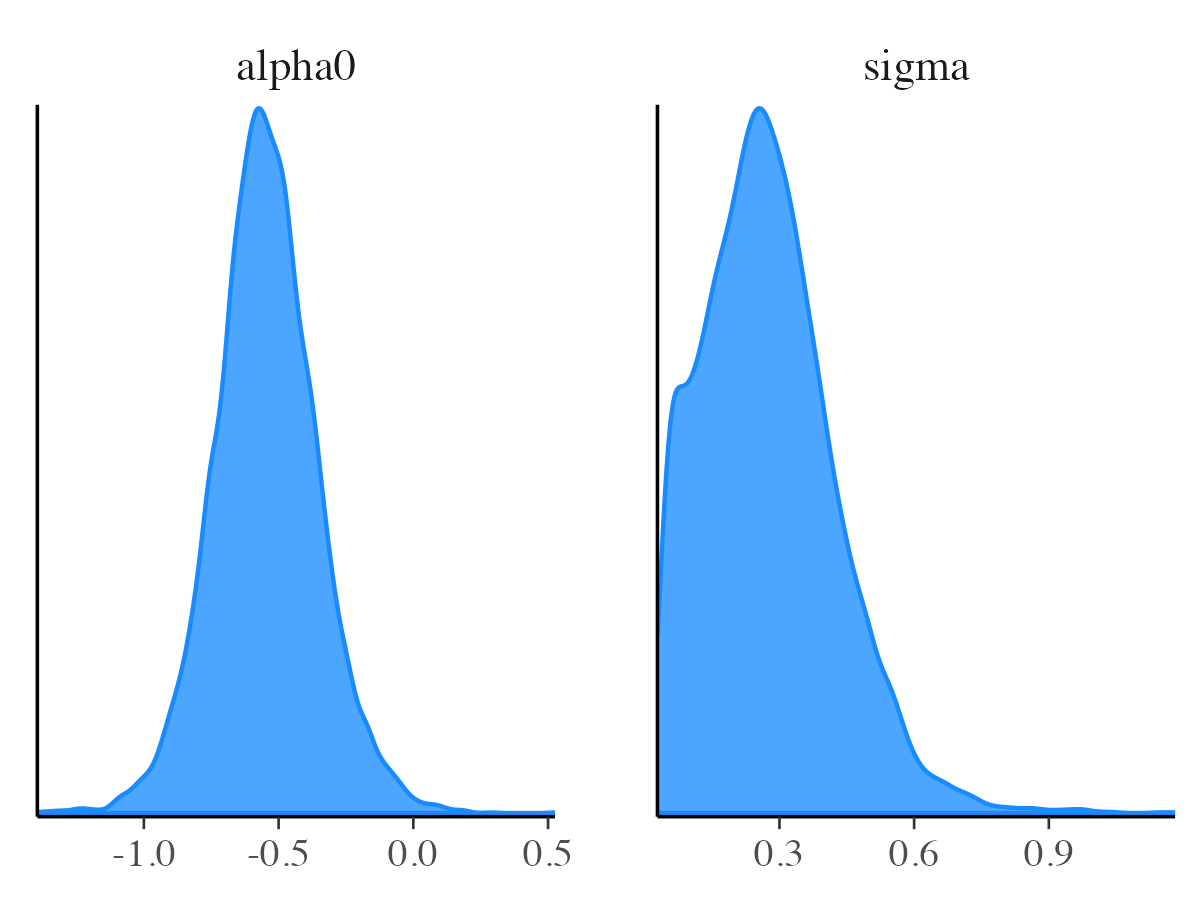
mcmc_dens_overlay
Similar to mcmc_hist_by_chain, the
mcmc_dens_overlay function distinguishes between Markov
chains; however, rather than showing them in separate panels, it
overlays their density estimates in a single plot.
mcmc_dens_overlay(posterior_samples, pars = c("alpha0", "sigma"))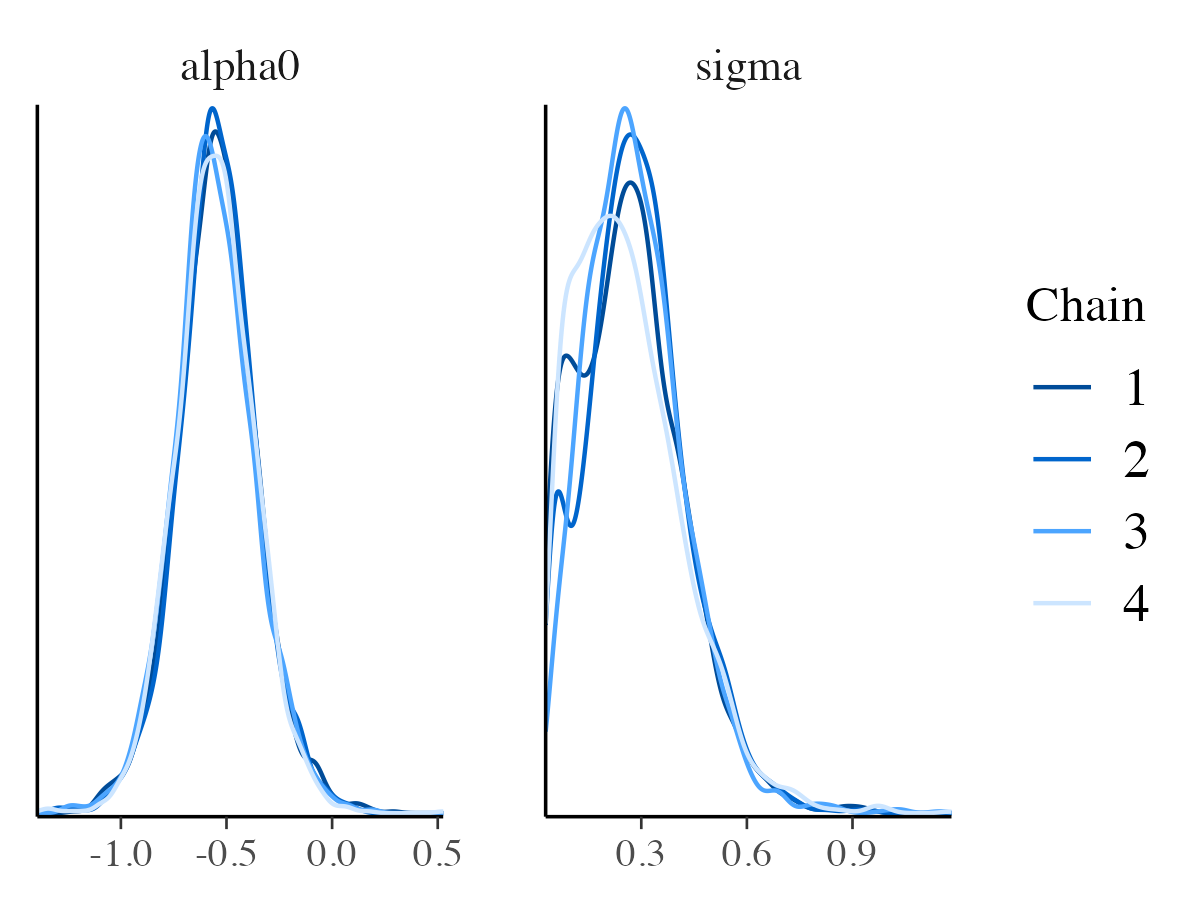
mcmc_violin
The mcmc_violin function visualizes the density
estimates for each chain using violin plots and adds horizontal lines at
quantiles specified by the user.
color_scheme_set("teal")
mcmc_violin(posterior_samples, pars = c("alpha0", "alpha12"))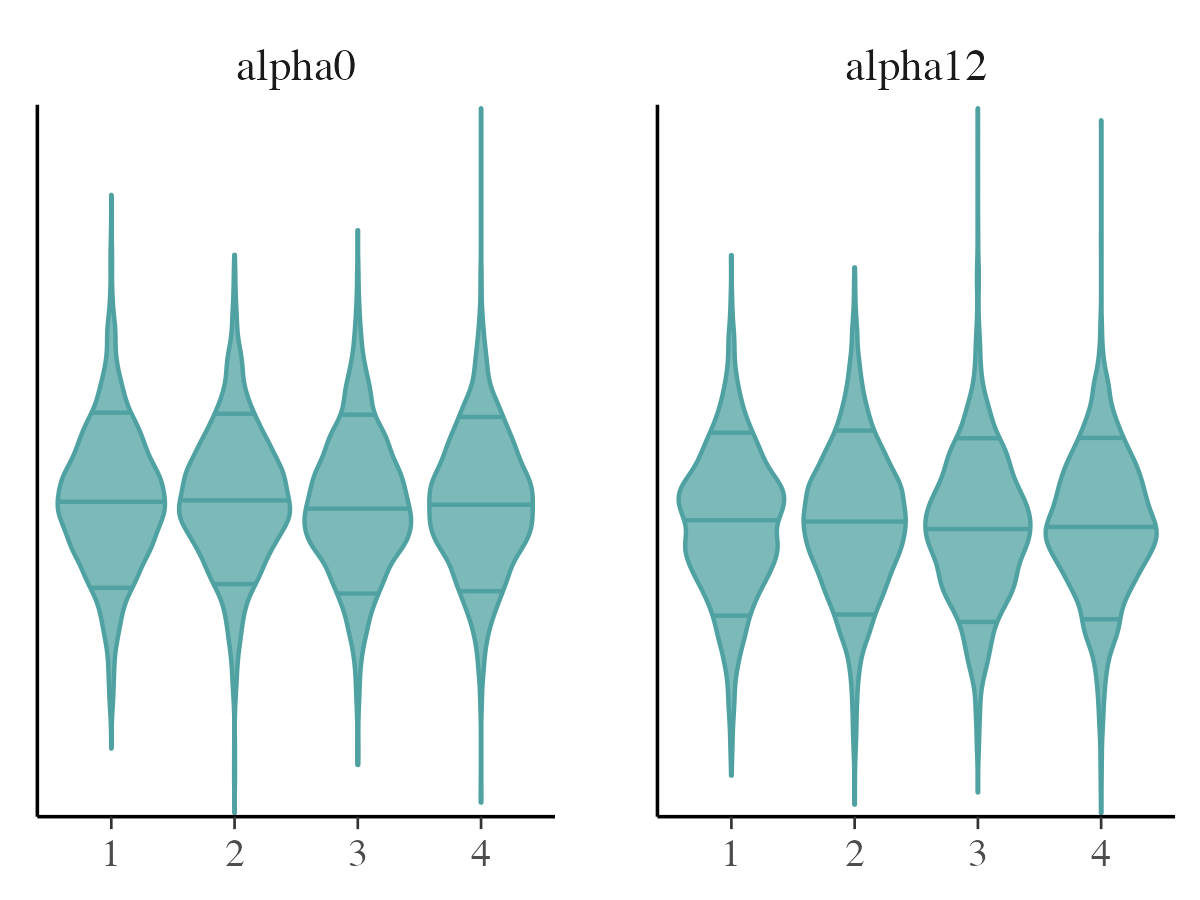
Bivariate plots
A range of functions can be used to visualize bivariate marginal posterior distributions. Several of these functions allow optional parameters to incorporate MCMC diagnostic details into the plots. The extended diagnostic features are explained in a separate vignette focused on visual MCMC diagnostics. The examples here replicate some examples but for complete reference check the original documentation
mcmc_scatter
The mcmc_scatter function generates a basic scatter plot
showing the relationship between two parameters.
color_scheme_set("orange")
mcmc_scatter(posterior_samples,
pars = c("alpha0", "alpha12"),
size = 1.5,
alpha = 0.5)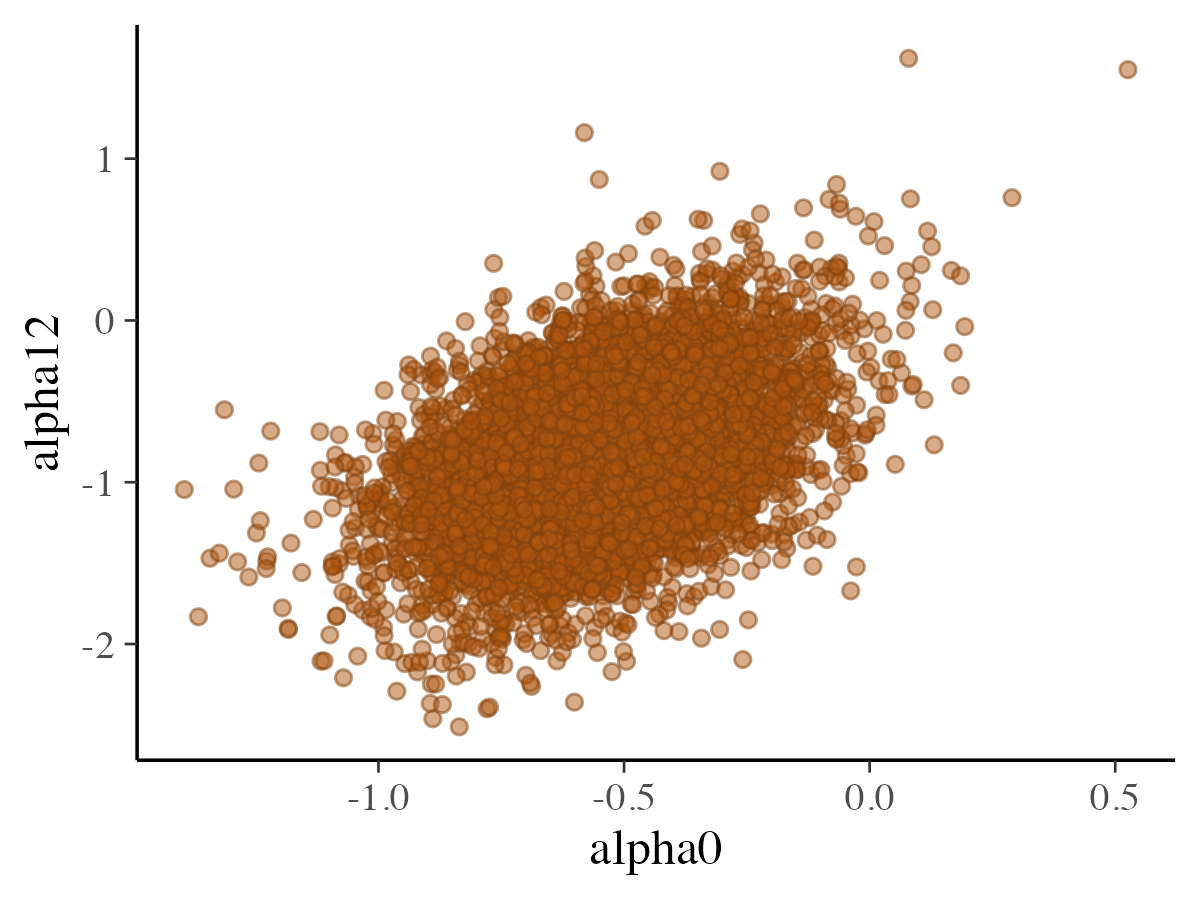
mcmc_hex
The mcmc_hex function produces a comparable plot using
hexagonal bins, which helps reduce overplotting when many points
overlap.
color_scheme_set("gray")
# requires hexbin package
if (requireNamespace("hexbin", quietly = TRUE)) {
mcmc_hex(posterior_samples, pars = c("alpha0", "alpha12"))
}
mcmc_pairs
In addition to mcmc_scatter and mcmc_hex,
the bayesplot package also includes the mcmc_pairs
function, which allows users to create pairs plots for exploring
relationships among multiple parameters.
color_scheme_set("purple")
# requires hexbin package
mcmc_pairs(posterior_samples,
pars = c("alpha0", "alpha1","alpha12"),
off_diag_args = list(size = 1.5))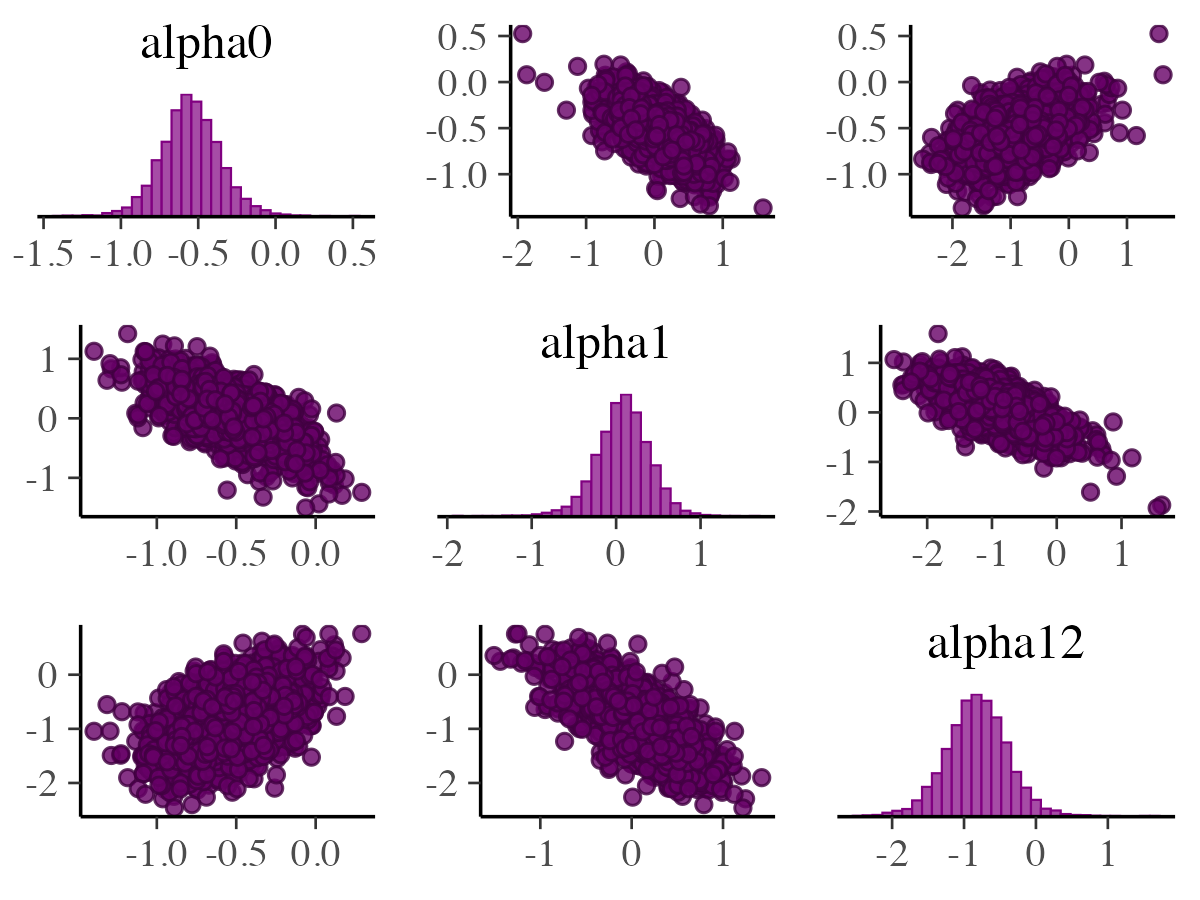
Univariate marginal posterior distributions appear along the diagonal
as histograms by default, but this can be switched to density plots
using diag_fun = "dens". The bivariate relationships are
shown above and below the diagonal using scatter plots, though these can
be replaced with hex-bin plots by setting
off_diag_fun = "hex". By default, mcmc_pairs
splits the Markov chains—showing half above the diagonal and the rest
below if the number of chains is even. Additional options for
customizing how chains are divided across these plots are available
through the condition argument, although these settings are
primarily useful when including MCMC diagnostic information. Further
details on that are provided in the official vignette from
bayesplot documentation named Visual
MCMC Diagnostics vignette.
Trace plots
Trace plots display Markov chains smaples, allowing users to inspect
how parameter values evolve over iterations. This section presents the
standard trace plots available in the bayesplot
package.
mcmc_trace
The mcmc_trace function generates basic trace plots to
visualize the sampling paths of parameters across iterations.
color_scheme_set("blue")
mcmc_trace(posterior_samples, pars = c("alpha0", "sigma"))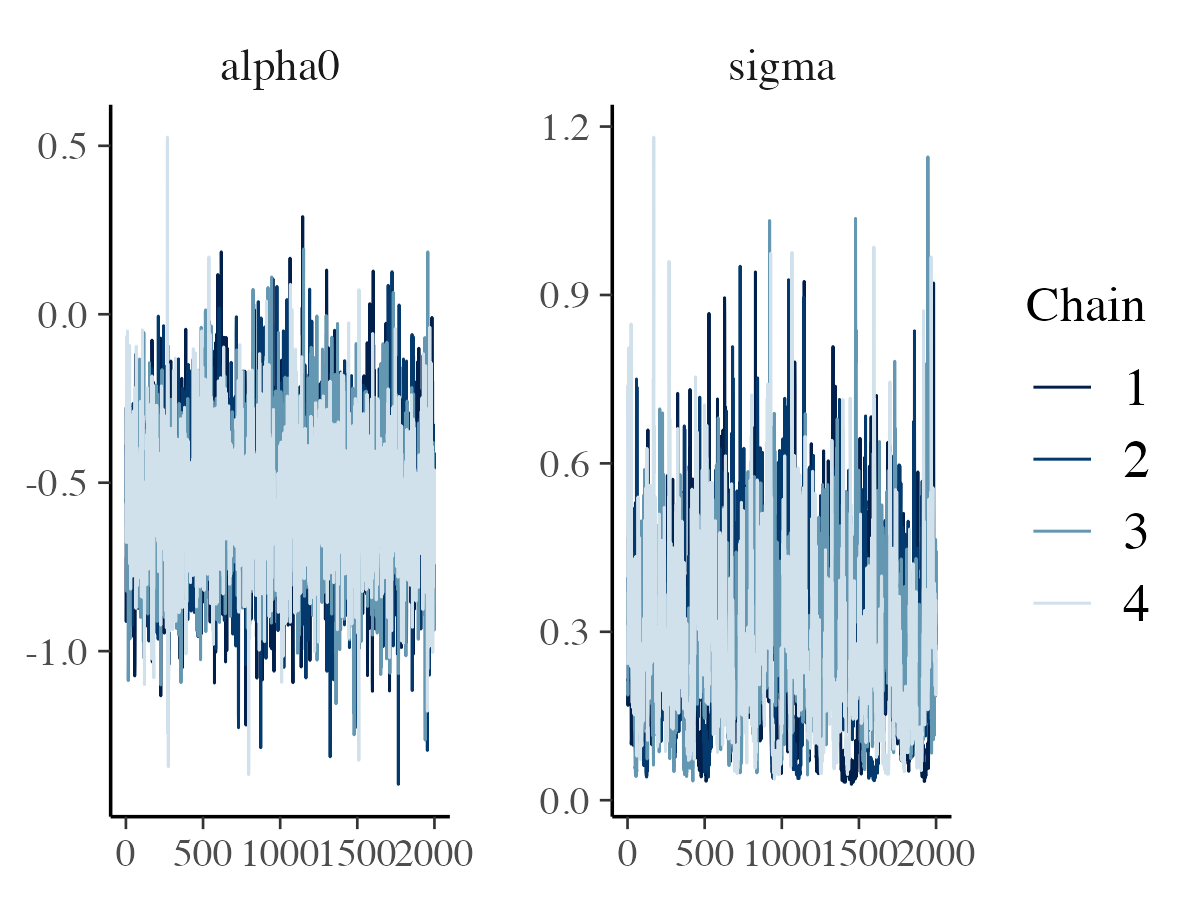
If the differences between chains are difficult to distinguish, a mixed color scheme can be used to improve clarity. For example:
color_scheme_set("mix-blue-red")
mcmc_trace(posterior_samples, pars = c("alpha0", "sigma"),
facet_args = list(ncol = 1, strip.position = "left"))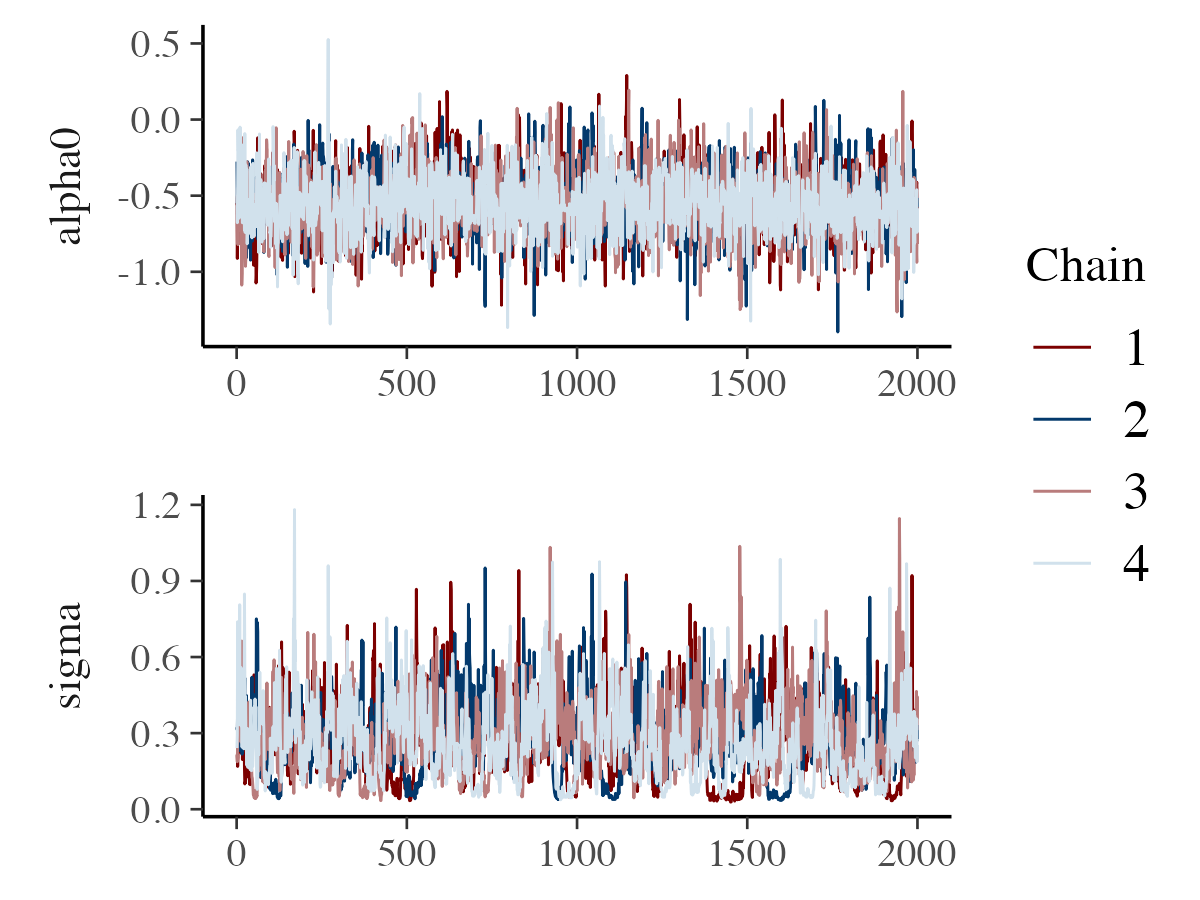
The example above also demonstrates the facet_args
argument, which allows you to pass options to ggplot2’s
facet_wrap. Setting ncol = 1 stacks the trace
plots in a single column, and strip.position = "left" moves
the facet labels to the y-axis rather than displaying them above each
plot.
mcmc_trace_highlight
The mcmc_trace_highlight function displays points
instead of lines and lowers the opacity of all chains except one, which
is emphasized using the highlight argument.
color_scheme_set("viridis")
mcmc_trace_highlight(posterior_samples, pars = c( "sigma"),
highlight = 1)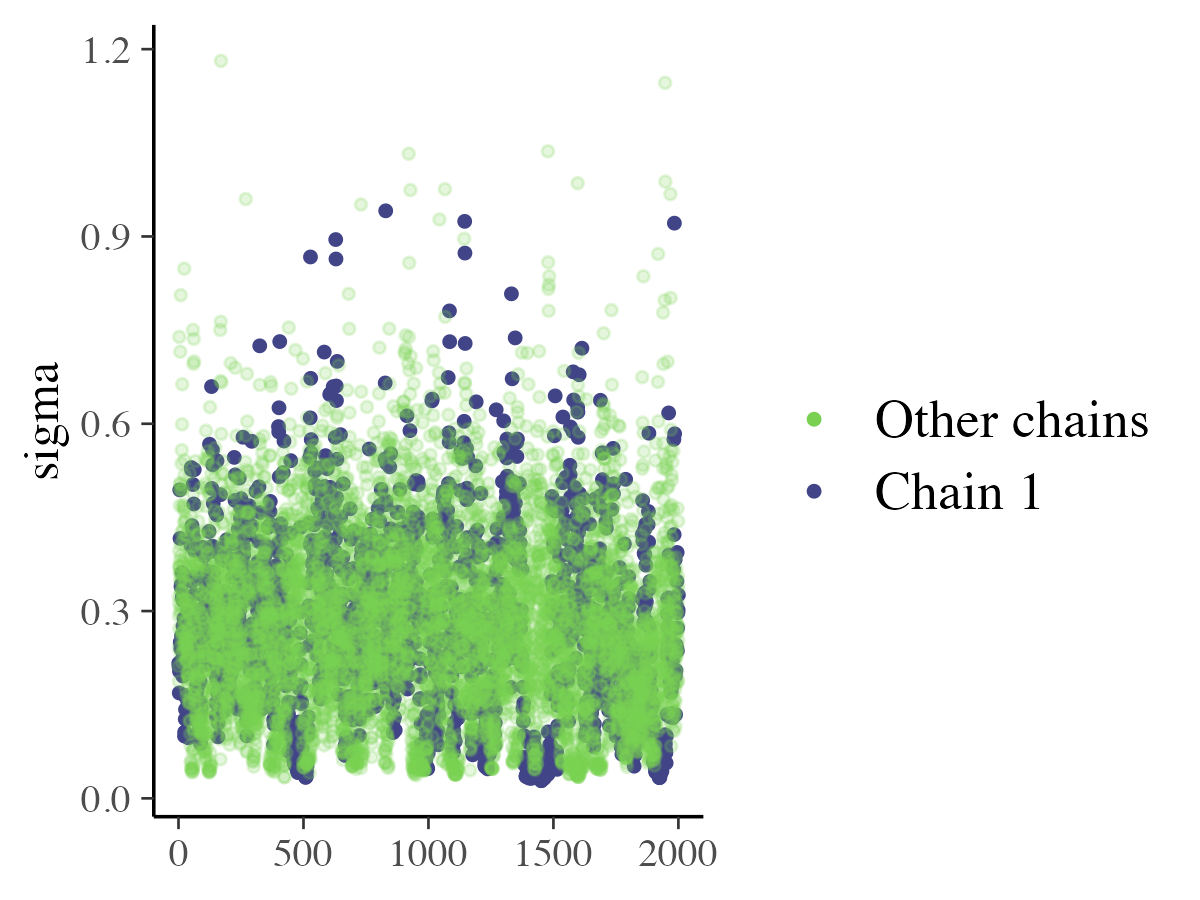
While trace plots focus on how the Markov chains evolve over
iterations, mcmc_rank_hist() examines how well the chains
mix by comparing the distribution of their ranks. Ideally, the histogram
should be roughly uniform, indicating good mixing across chains. See
Vehtari et al. (2021) for more background.
mcmc_rank_hist(posterior_samples, pars = c("sigma"))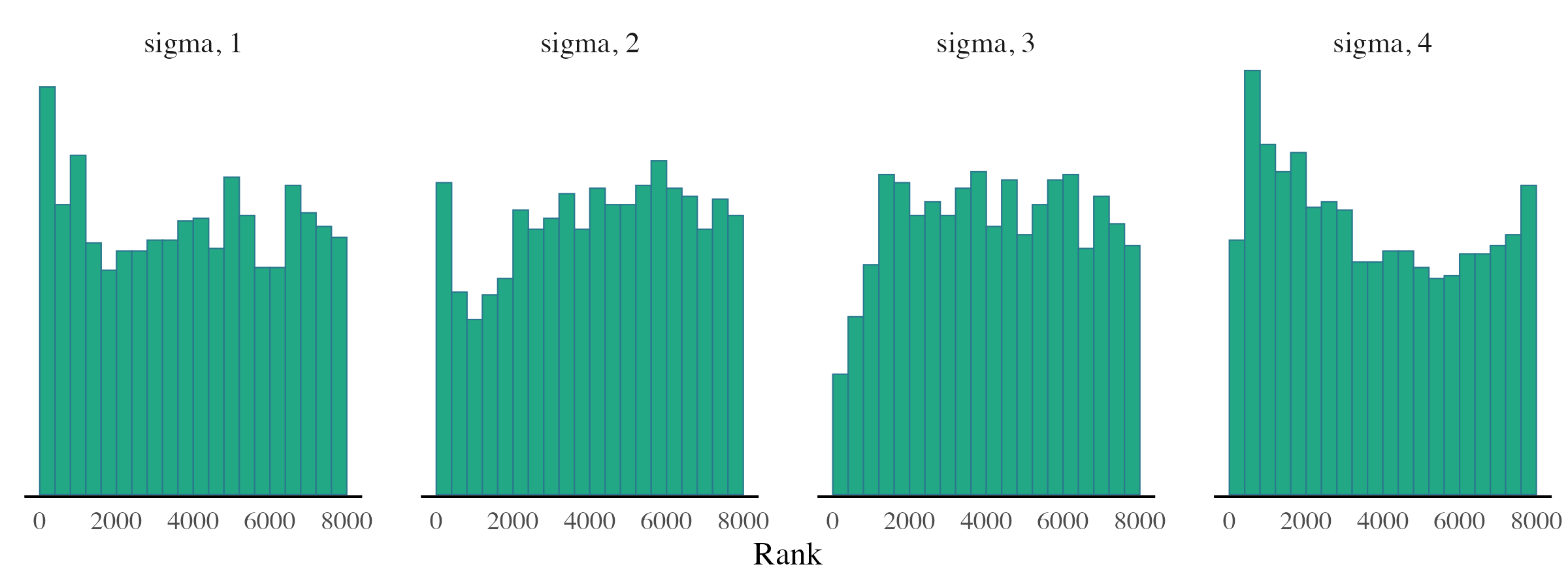
Additionally, mcmc_rank_ecdf() plots the empirical
cumulative distribution functions (ECDFs) of the ranks along with
simultaneous confidence bands. These bands have a coverage probability
controlled by the prob argument, meaning they are designed
to fully contain all rank ECDFs with probability prob. When
plot_diff = TRUE, the plot also shows the difference
between the observed rank ECDFs and the theoretical uniform distribution
expected if all samples came from the same distribution. For more
details, see Säilynoja et al. (2022). The function arguments are similar
to mcmc_rank_hist.
References
Aki Vehtari, Andrew Gelman, Daniel Simpson, Bob Carpenter, Paul-Christian Bürkner “Rank-Normalization, Folding, and Localization: An Improved R-hat for Assessing Convergence of MCMC (with Discussion),” Bayesian Analysis, Bayesian Anal. 16(2), 667-718, (June 2021)
Säilynoja, T., Bürkner, PC. & Vehtari, A. Graphical test for discrete uniformity and its applications in goodness-of-fit evaluation and multiple sample comparison. Stat Comput 32, 32 (2022). https://doi.org/10.1007/s11222-022-10090-6
Gabry, J., and Goodrich, B. (2017). rstanarm: Bayesian Applied Regression Modeling via Stan. R package version 2.15.3. https://mc-stan.org/rstanarm/, https://CRAN.R-project.org/package=rstanarm
Gabry, J., Simpson, D., Vehtari, A., Betancourt, M. and Gelman, A. (2019), Visualization in Bayesian workflow. J. R. Stat. Soc. A, 182: 389-402. :10.1111/rssa.12378. (journal version, arXiv preprint, code on GitHub)
Gelman, A., Carlin, J. B., Stern, H. S., Dunson, D. B., Vehtari, A., and Rubin, D. B. (2013). Bayesian Data Analysis. Chapman & Hall/CRC Press, London, third edition.
Stan Development Team. (2024). Stan Reference Manual, Version 2.36. https://mc-stan.org
Stan Development Team. (2024). bayesplot: Plotting for MCMC draws. https://mc-stan.org/bayesplot/articles/plotting-mcmc-draws.html